In Silico Strategies for Therapeutic Discovery: Predicting Molecular Properties, Targets, and Metabolic Interactions

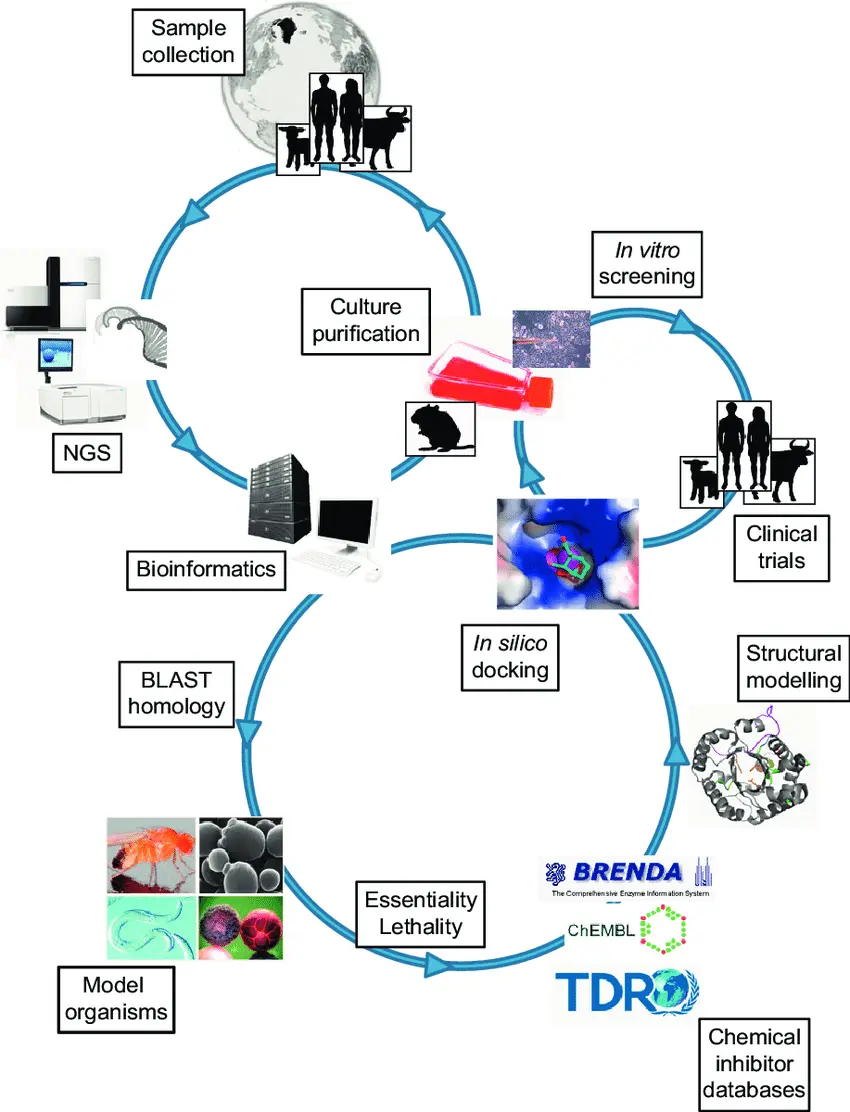
The journey to bring a new therapeutic molecule to patients is long, expensive, and full of challenges. Traditionally, scientists have relied heavily on laboratory experiments and clinical trials to test thousands of compounds, but this process can take over a decade and cost billions of dollars. Today, in silico approaches meaning computer-based methods are changing the way researchers design and optimize bioactive molecules.
In silico tools help predict how small molecules will behave in the body by providing valuable information about their biological properties, likely targets, and metabolic interactions. These predictions allow researchers to make better decisions early in the discovery process, save time and money, and reduce the number of compounds that fail during clinical testing.
What Are In Silico Strategies?
In silico strategies refer to the use of computer modeling, simulations, and data analysis to predict or simulate the behavior of molecules. These methods combine structural biology, chemistry, bioinformatics, and big data to generate insights that once required extensive lab work.

Predicting biological and physicochemical properties of small molecules.
Predicting biological and physicochemical properties of small molecules.
Simulating how molecules bind to receptors or enzymes.

Assessing possible toxicities and interactions with metabolic enzymes and transporters.

Predicting how compounds will be absorbed, distributed, metabolized, and excreted in the body (ADME).

Why Predict Molecular Properties?
Before any molecule becomes a successful therapeutic agent, it must have suitable properties that allow it to reach its target and have the desired effect.
In silico methods can predict key features such as:
Solubility
Whether a molecule can dissolve in bodily fluids. Poor solubility can mean poor absorption.
Lipophilicity (LogP)
Indicates how easily a molecule can pass through cell membranes.
Stability
Predicts if a molecule will remain intact long enough to be effective.
Permeability
Shows how well a molecule can cross biological barriers like the gut lining or blood-brain barrier.
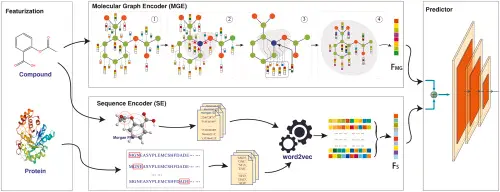
Predicting and Validating Biological Targets
Finding the right target is crucial for developing an effective therapeutic molecule. A target can be a protein, enzyme, receptor, or even a specific part of a cell that the molecule will interact with to produce a desired biological response.
In silico target prediction combines several techniques:
Virtual Screening
Large libraries of compounds are screened computationally against potential biological targets to identify promising hits.
This method uses advanced algorithms to identify which compounds are most likely to bind effectively to a target’s active site based on their structural and physicochemical properties. By narrowing down thousands or even millions of potential candidates to a smaller, more promising list, virtual screening helps researchers prioritize which molecules should move forward for laboratory testing and optimization. This approach saves significant time and resources, improves hit rates, and increases the likelihood of discovering bioactive compounds with desirable therapeutic effects.
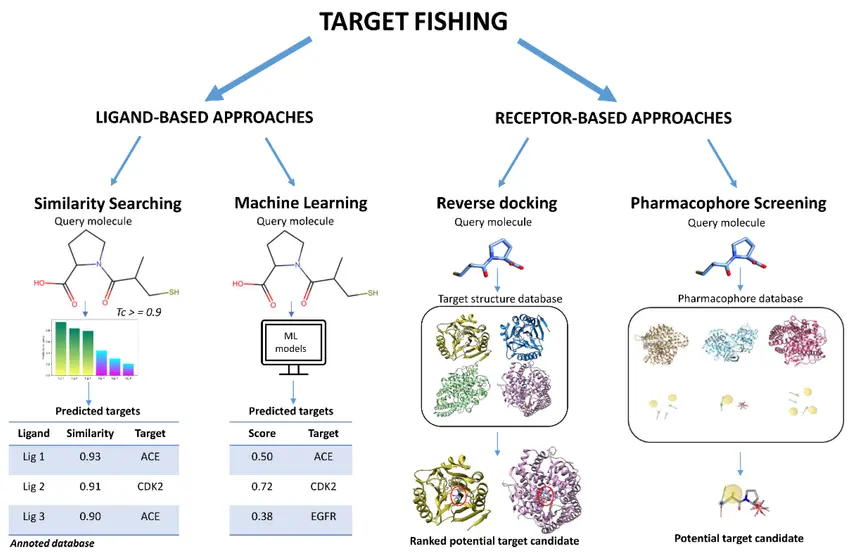
Target Fishing
Target fishing is an important in silico strategy used to predict and validate potential biological targets for small molecules. This approach involves using computational algorithms and large biological databases to identify which proteins, receptors, or enzymes a compound is most likely to bind to. By comparing the chemical structure of a molecule to known ligands and interaction patterns, researchers can quickly generate hypotheses about its possible mechanisms of action. Target fishing helps uncover unexpected off-target effects or new therapeutic opportunities, saving valuable time and resources in the laboratory. This prediction step is crucial for prioritizing which targets to test experimentally, ensuring that the most promising interactions are investigated further in the drug discovery pipeline.
Reverse docking
Reverse docking is an important in silico strategy used to predict and validate potential biological targets for a small molecule when the target is unknown or when researchers want to discover additional off-target interactions. Unlike traditional docking, which tests multiple molecules against one known protein, reverse docking works the other way around: it screens a single compound against a large database of protein structures to identify possible binding sites across different targets. This approach helps scientists uncover unexpected protein interactions, understand potential side effects, and find opportunities for drug repurposing. By using reverse docking, researchers can generate valuable hypotheses about a molecule’s mechanism of action and prioritize experimental studies on the most promising target candidates, saving significant time and resources during early discovery.
Simulating Ligand-Receptor Interactions
Prediction of Cytochrome P450 Substrates
One of the biggest challenges in developing safe and effective therapeutic molecules is understanding how they are metabolized in the human body. Metabolism determines how long a compound stays active, how it is broken down, and how it is eliminated. Poor metabolic stability can lead to low effectiveness, toxic by-products, or unwanted interactions with other substances.
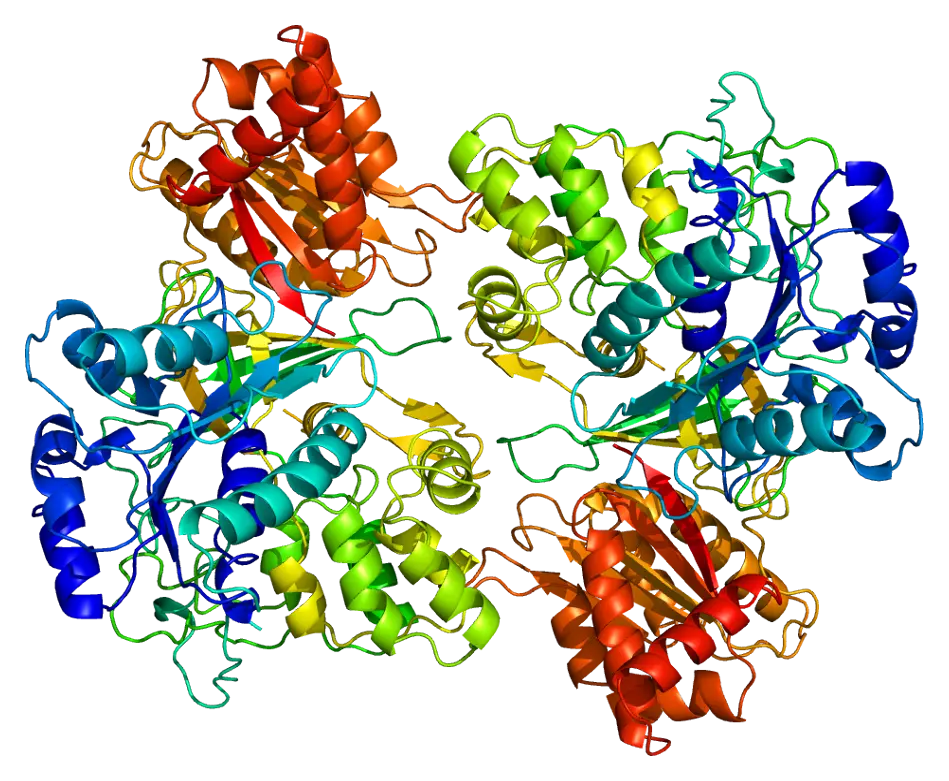
The Cytochrome P450 (CYP450) enzyme family plays a central role in this process. In humans, these enzymes are responsible for metabolizing around 70–80% of all small molecules used as therapeutic agents. Therefore, predicting how a new molecule will interact with CYP450s is essential for assessing its safety, efficacy, and potential for drug–drug interactions.
Benefits and Limitations
In silico methods offer significant advantages: they reduce experimental costs, shorten timelines, and limit the need for animal testing. However, these techniques rely on the availability and quality of experimental data to build reliable models. While they provide valuable predictions, they are most effective when combined with in vitro and in vivo studies to validate findings.